Testing alt.tbo with Atl3_bis 20171112a
import numpy as np
import matplotlib.pyplot as plt
import math
import os
import bim as USTk
for filename in os.listdir("./"):
if filename.endswith(".DAT"):
DATFile = os.path.join("", filename).split(".")[0]
if not os.path.isfile(DATFile+".npz"):
Arf = USTk.CreateUsPack(DATFile+".DAT")
NP1.DAT

OP1.DAT

NP3.DAT

NP4.DAT

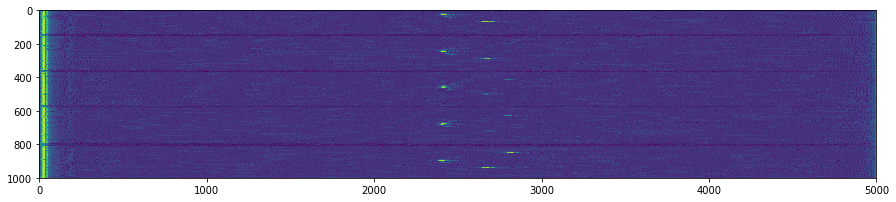
NP2.DAT

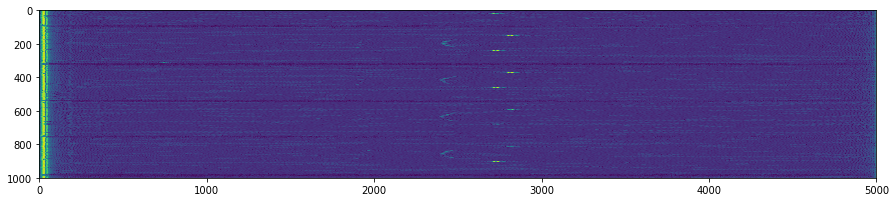
OP3.DAT

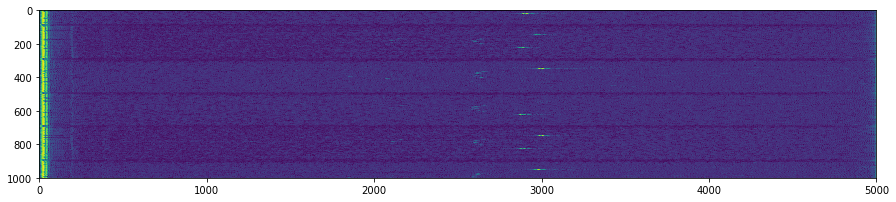
## Creates the npz files from the DAT files, with reshaped signal, and filtered images
# USTk.CreateUsPack(DATFile+".DAT")
for filename in os.listdir("./"):
if filename.endswith(".npz"):
print filename
DATFile = os.path.join("", filename).split(".")[0]
DATA = np.load(DATFile+".npz")
Image, rawSignal = DATA['arr_1'], DATA['arr_0']
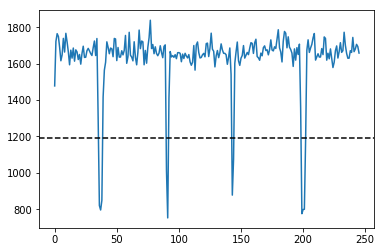
IMGs, GoodV, tmp = USTk.CreateRawImgs (Image,5)
## Cell 2
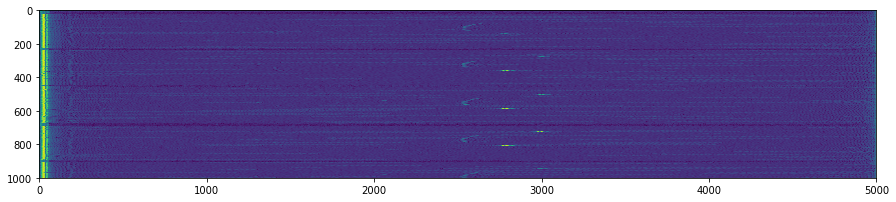
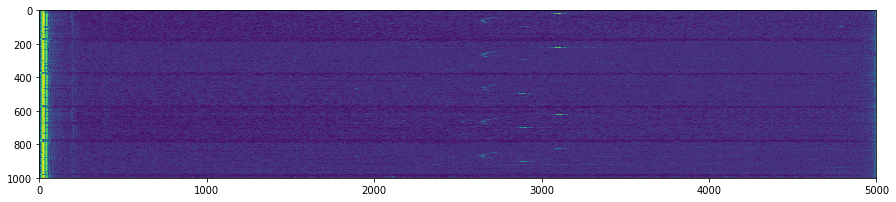
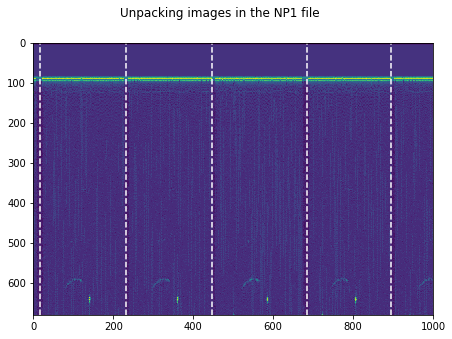
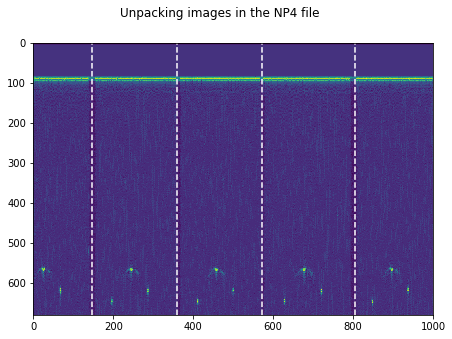
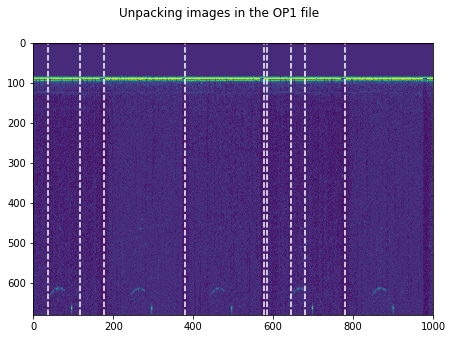
plt.figure(figsize=(15,5))
plt.imshow(np.transpose(tmp))
for k in GoodV:
plt.axvline(k, color='w', linestyle='--')
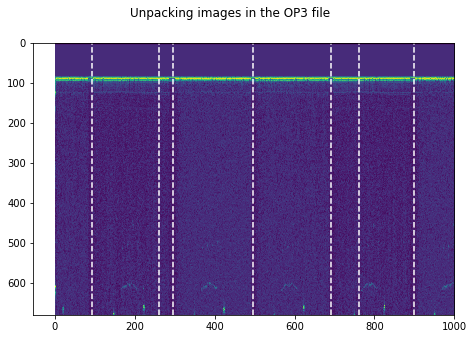
plt.suptitle('Unpacking images in the '+DATFile+' file')
plt.savefig('raw/Unpacking_'+DATFile+".jpg", bbox_inches='tight')
plt.show()
## CEll 3
N = len(IMGs)
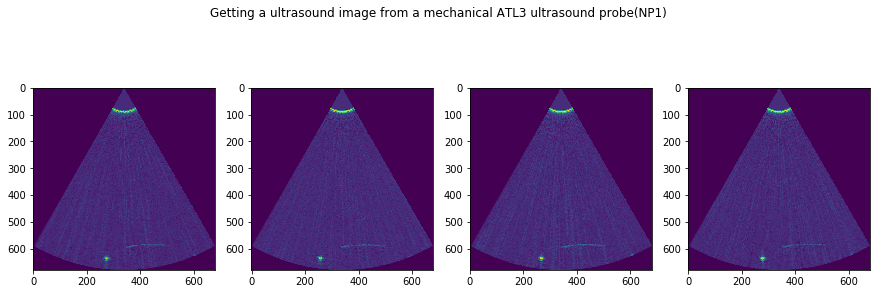
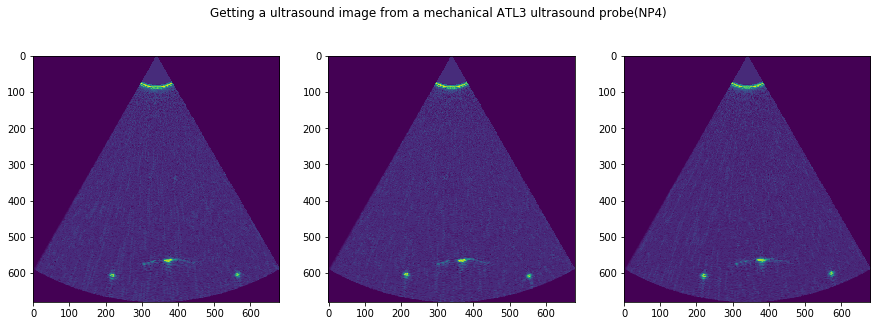
f, AX = plt.subplots(1, N, figsize=(15,5))
for i in range(N):
tmp = USTk.CreateSC(IMGs[i])
AX[i].imshow(tmp[4])
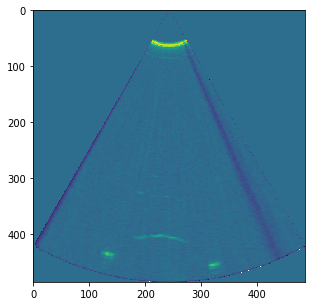
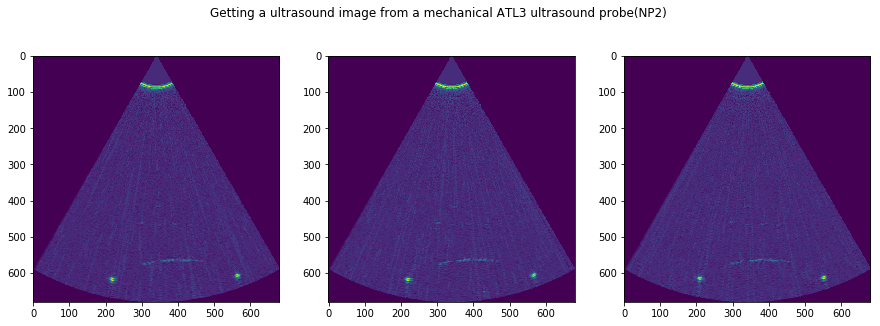
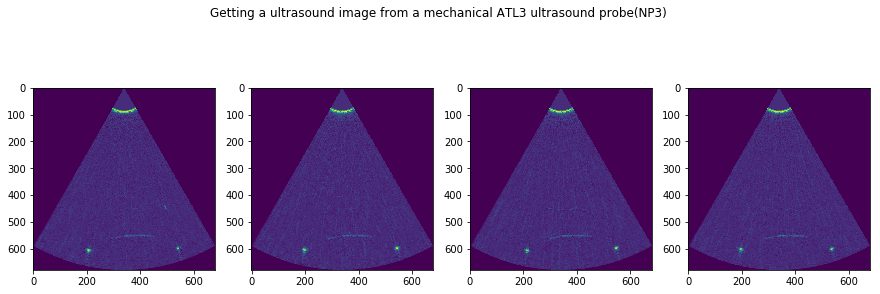
plt.suptitle('Getting a ultrasound image from a mechanical ATL3 ultrasound probe('+str(DATFile)+')')
plt.savefig('Images_'+DATFile+".jpg", bbox_inches='tight')
plt.show()
NP1.npz
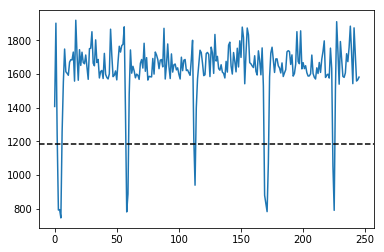
NP2.npz
NP3.npz

NP4.npz
OP1.npz
/usr/local/lib/python2.7/dist-packages/numpy/lib/function_base.py:1110: RuntimeWarning: Mean of empty slice.
avg = a.mean(axis)
/usr/local/lib/python2.7/dist-packages/numpy/core/_methods.py:80: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
OP3.npz
DATFile = "NP2"
DATA = np.load(DATFile+".npz")
Image, rawSignal = DATA['arr_1'], DATA['arr_0']
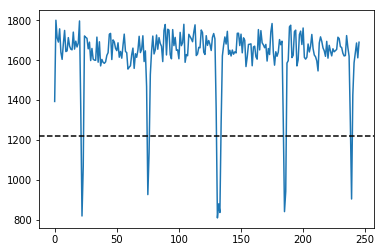
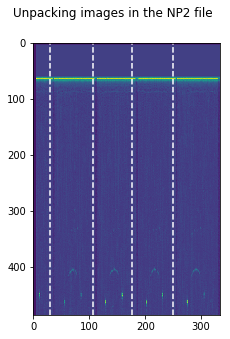
IMGs, GoodV, tmp = USTk.CreateRawImgsLR (Image,7,3)
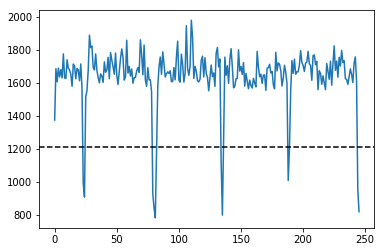
## Cell 2
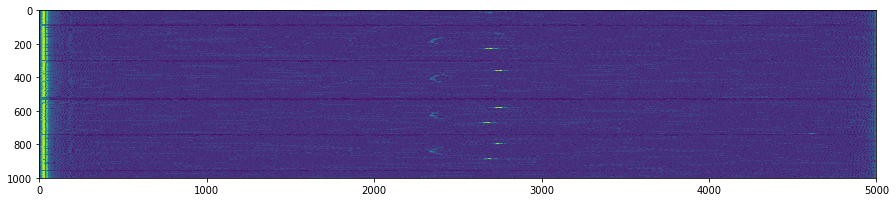
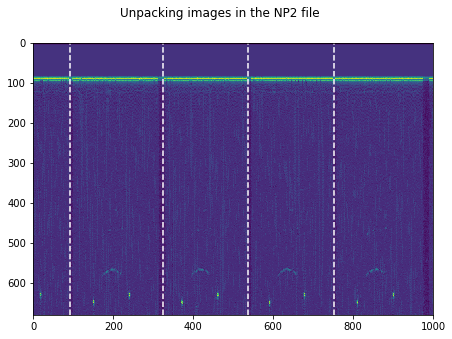
plt.figure(figsize=(15,5))
plt.imshow(np.transpose(tmp))
for k in GoodV:
plt.axvline(k, color='w', linestyle='--')
plt.suptitle('Unpacking images in the '+DATFile+' file')
plt.savefig('Unpacking_'+DATFile+".jpg", bbox_inches='tight')
plt.show()
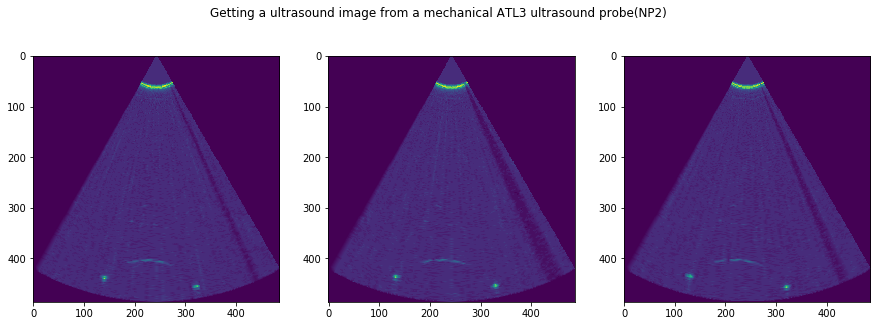
## CEll 3
N = len(IMGs)
f, AX = plt.subplots(1, N, figsize=(15,5))
for i in range(N):
tmp = USTk.CreateSC(IMGs[i])
AX[i].imshow(tmp[4])
plt.suptitle('Getting a ultrasound image from a mechanical ATL3 ultrasound probe('+str(DATFile)+')')
plt.savefig('ImagesIn_'+DATFile+".jpg", bbox_inches='tight')
plt.show()
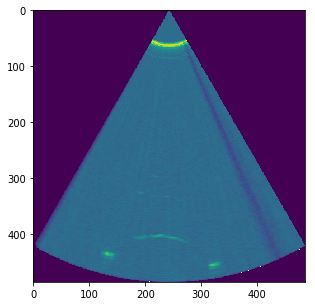
Res = USTk.CreateSC(IMGs[0])[4]
for i in range(N-1):
tmp = USTk.CreateSC(IMGs[i+1])[4]
Res += tmp
plt.figure(figsize=(15,5))
plt.imshow(np.asarray(np.sqrt(Res)))
plt.show()
/usr/local/lib/python2.7/dist-packages/ipykernel/__main__.py:2: RuntimeWarning: invalid value encountered in sqrt
from ipykernel import kernelapp as app
len(Res)
len(Res)
486
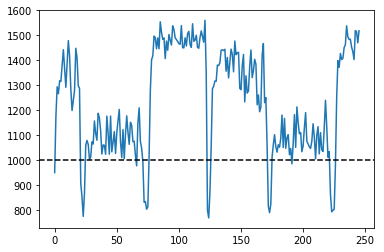
for k in range(len(Res)):
for i in range(len(Res)):
if Res[i][k] == 0:
Res[i][k] = Res[20][243]
plt.figure(figsize=(15,5))
plt.imshow(np.asarray(np.sqrt(Res)))
plt.show()
/usr/local/lib/python2.7/dist-packages/ipykernel/__main__.py:2: RuntimeWarning: invalid value encountered in sqrt
from ipykernel import kernelapp as app