20180516a - JSON Client - version e from un0rick
import numpy as np
import matplotlib.pyplot as plt
from scipy import signal
from scipy.interpolate import griddata
import math
from scipy.signal import decimate, convolve
import json
import re
import glob, os
import sys
import pyexiv2
import os
Creating the set of acquisitions
def MetaDataImg(Modules,Experiment,Category,Description):
Imgs = []
for dirpath, dirnames, filenames in os.walk("."):
for filename in [f for f in filenames if ( f.endswith(".jpg") or f.endswith(".png") )]:
Imgs.append( os.path.join(dirpath, filename) )
for FileName in Imgs:
edit = 0
metadata = pyexiv2.ImageMetadata(FileName)
try:
metadata.read()
except IOError:
print "Not an image"
else:
metadata['Exif.Image.Software'] = Modules
metadata['Exif.Image.Make'] = Experiment
metadata['Exif.Photo.MakerNote'] = Category
metadata['Exif.Image.ImageDescription'] = Description
metadata.write()
print FileName, "done"
class us_json:
IDLine = []
TT1 = []
TT2 = []
tmp = []
tdac = []
FFT_x = []
FFT_y = []
FFT_filtered = []
LengthT = 0
Nacq = 0
Raw = []
Signal = []
SignalFiltered = []
Registers = {}
t= []
fPiezo = 3.5
f = 0
firmware_md5 = ""
experiment = ""
Nacq = 0
len_acq = 0
len_line= 0
N = 0
V = 0
single = 0
processed = False
iD = 0
def JSONprocessing(self,path):
IDLine = []
TT1 = []
TT2 = []
tmp = []
tdac = []
with open(path) as json_data:
DATA = {}
d = json.load(json_data)
json_data.close()
A = d["data"][1:]
print d.keys()
if (A[0]) > 128:
print "first"
for i in range(len(A)/2-1):
value = 128*(A[2*i+0]&0b0000111) + A[2*i+1] - 512
IDLine.append((A[2*i+0]&0b11110000)/16)
TT1.append( (A[2*i+0] & 0b00001000) / 0b1000)
TT2.append( (A[2*i+0] & 0b00010000) / 0b10000)
tmp.append( 2.0*value/512.0 )
else:
print "second"
for i in range(len(A)/2-1):
value = 128*(A[2*i+1]&0b111) + A[2*i+2] - 512
IDLine.append((A[2*i+1]&0b11110000)/16)
TT1.append( (A[2*i+1] & 0b00001000) / 0b1000)
TT2.append( (A[2*i+1] & 0b00010000) / 0b10000)
tmp.append( 2.0*value/512.0 )
self.f = float(64/((1.0+int( d["registers"]["237"] ) )))
t = [ 1.0*x/self.f for x in range(len(tmp))]
self.t = t
self.LengthT = len(t)
self.FFT_x = [ X*self.f / (self.LengthT) for X in range(self.LengthT)]
self.FFT_y = np.fft.fft(tmp)
self.FFT_filtered = np.fft.fft(tmp)
for k in range (self.LengthT/2 + 1):
if k < (self.LengthT * self.fPiezo * 0.5 / self.f):
self.FFT_filtered[k] = 0
self.FFT_filtered[-k] = 0
if k > (self.LengthT * self.fPiezo *1.5 / self.f):
self.FFT_filtered[k] = 0
self.FFT_filtered[-k] = 0
self.SignalFiltered = np.real(np.fft.ifft(self.FFT_filtered))
self.TT1 = TT1
self.TT2 = TT2
self.Nacq = d["registers"][str(0xEC)]
self.len_acq = len(self.t)
self.len_line = self.len_acq/self.Nacq
self.Registers = d["registers"]
REG = [int(x) for x in d["registers"].keys() if int(x) < 100]
REG.sort()
dac = []
for k in REG:
dac.append(d["registers"][str(k)])
tdac = []
for pts in t[0:self.len_line]:
i = int(pts/5.0)
tdac.append(4.0*d["registers"][str(i+16)])
self.tdac = tdac
self.tmp = tmp
self.single = d["registers"][str(0XEB)]
self.t = t
self.IDLine = IDLine
self.firmware_md5 = d['firmware_md5']
self.experiment = d['experiment']
self.parameters = d['parameters']
self.iD = d['experiment']["id"]
self.N = d['N']
self.V = d['V']
self.processed = True
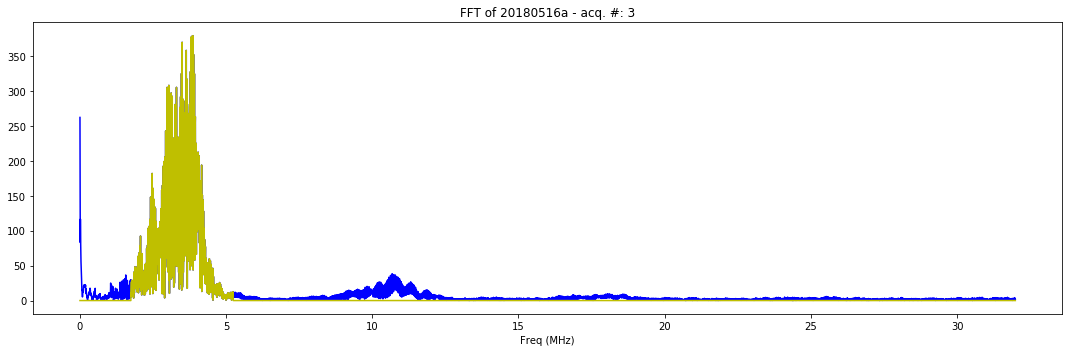
def mkFFT(self):
if self.processed:
plt.figure(figsize=(15,5))
plt.plot(self.FFT_x[1:self.LengthT/2], np.abs(self.FFT_y[1:self.LengthT/2]), 'b-')
plt.plot(self.FFT_x[1:self.LengthT/2], np.abs(self.FFT_filtered[1:self.LengthT/2]), 'y-')
plt.title( "FFT of "+self.iD + " - acq. #: "+ str(self.N))
plt.xlabel('Freq (MHz)')
plt.tight_layout()
FileName = "images/"+self.iD+"-"+str(self.N)+"-fft.jpg"
plt.savefig(FileName)
plt.show()
self.TagImage("matty,cletus",self.iD,"FFT","FFT of the of "+self.iD +" experiment. "+self.experiment["description"])
def mkImg(self):
if self.processed:
fig, ax1 = plt.subplots(figsize=(20,10))
ax2 = ax1.twinx()
ax2.plot(self.t[0:self.len_line], self.tdac[0:self.len_line], 'g-')
ax1.plot(self.t[0:self.len_line], self.tmp[0:self.len_line], 'b-')
plt.title( self.iD + " - acq. #: "+ str(self.N))
ax1.set_xlabel('Time (us)')
ax1.set_ylabel('Signal from ADC (V)', color='b')
ax2.set_ylabel('DAC output in mV (range 0 to 1V)', color='g')
plt.tight_layout()
FileName = "images/"+self.iD+"-"+str(self.N)+".jpg"
plt.savefig(FileName)
plt.show()
self.TagImage("matty,cletus",self.iD,"graph","Graph of "+self.iD +" experiment. "+self.experiment["description"])
def TagImage(self,Module,ID,Type,Description):
FileName = "images/"+self.iD+"-"+str(self.N)+".jpg"
metadata = pyexiv2.ImageMetadata(FileName)
try:
metadata.read()
except IOError:
print "Not an image"
else:
metadata['Exif.Image.Software'] = Module
metadata['Exif.Image.Make'] = ID
metadata['Exif.Photo.MakerNote'] = Type
metadata['Exif.Image.ImageDescription'] = Description
metadata.write()
def SaveNPZ(self):
NPZPath = "data/"+self.iD+"-"+str(self.N)+".npz"
np.savez(NPZPath, self)
print "Saved at "+NPZPath
y = us_json()
y.JSONprocessing("data/20180516a-1.json")
y.mkImg()
y.SaveNPZ()
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
first
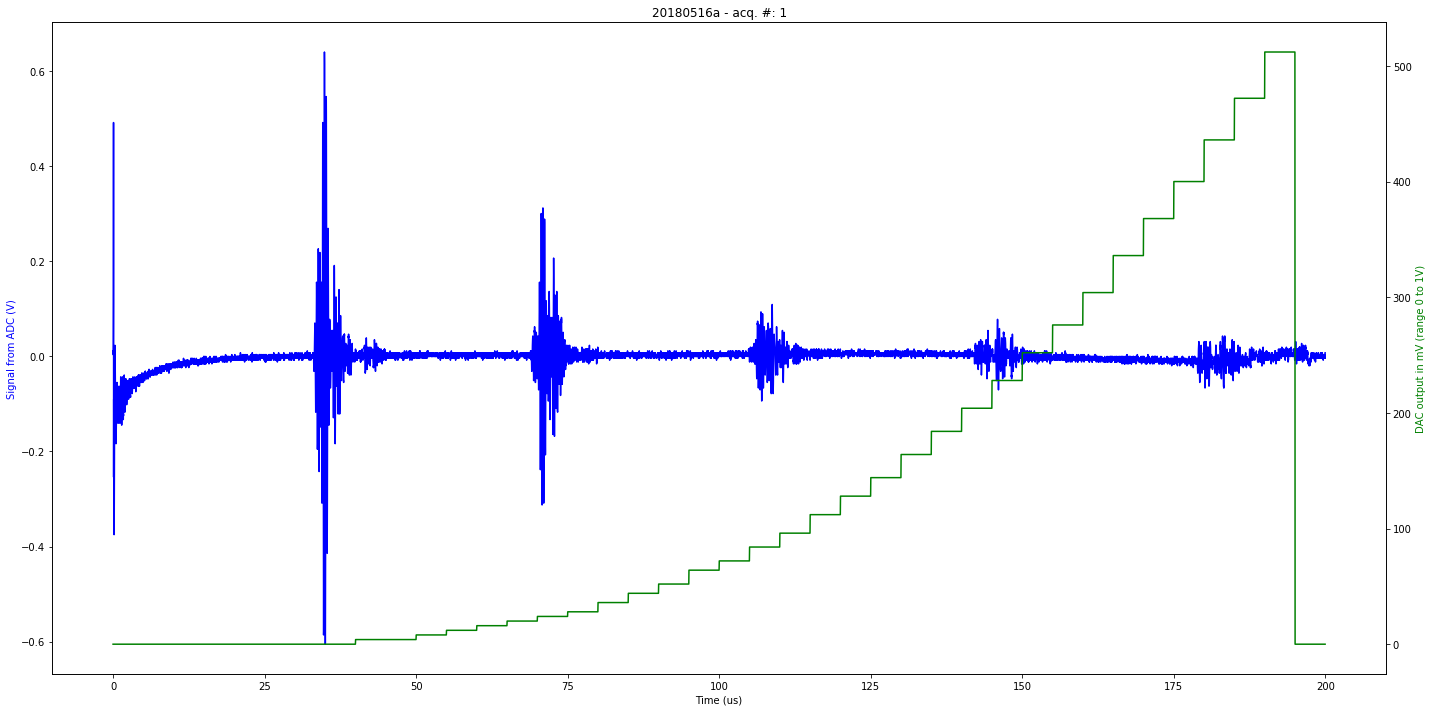
Saved at data/20180516a-1.npz
Checking the effects of a FFT cleaning
y.mkFFT()
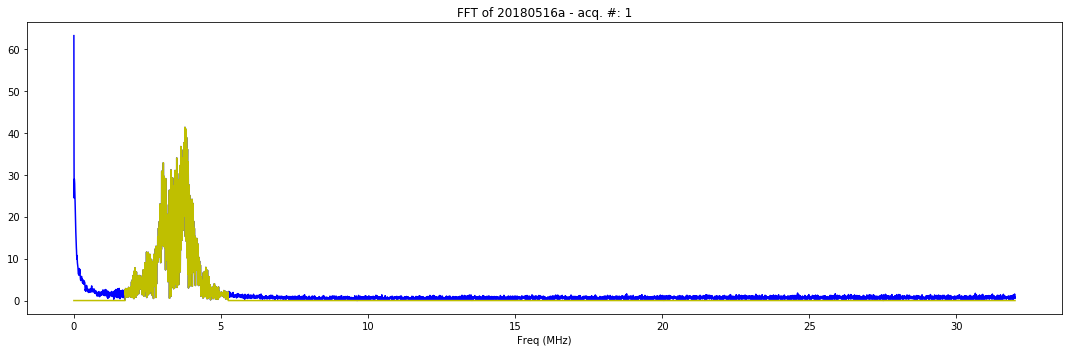
Checking the results of filtering
plt.figure(figsize=(15,5))
plt.plot(y.t[12799*178/200:12799*190/200], y.tmp[12799*178/200:12799*190/200], 'b-')
plt.plot(y.t[12799*178/200:12799*190/200], y.SignalFiltered[12799*178/200:12799*190/200], 'y-')
plt.title( "FFT of "+y.iD + " - acq. #: "+ str(y.N))
plt.xlabel('Freq (MHz)')
plt.tight_layout()
plt.show()
print len(y.t)
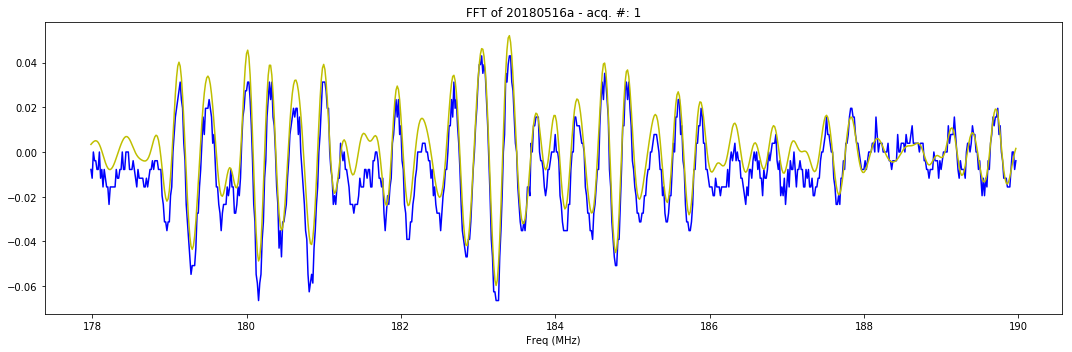
12799
DataSet = {}
for data in glob.glob("data/*.json"):
print data
x = us_json()
x.JSONprocessing(data)
x.mkImg()
x.SaveNPZ()
print x.Nacq
data/20180516a-3.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
first
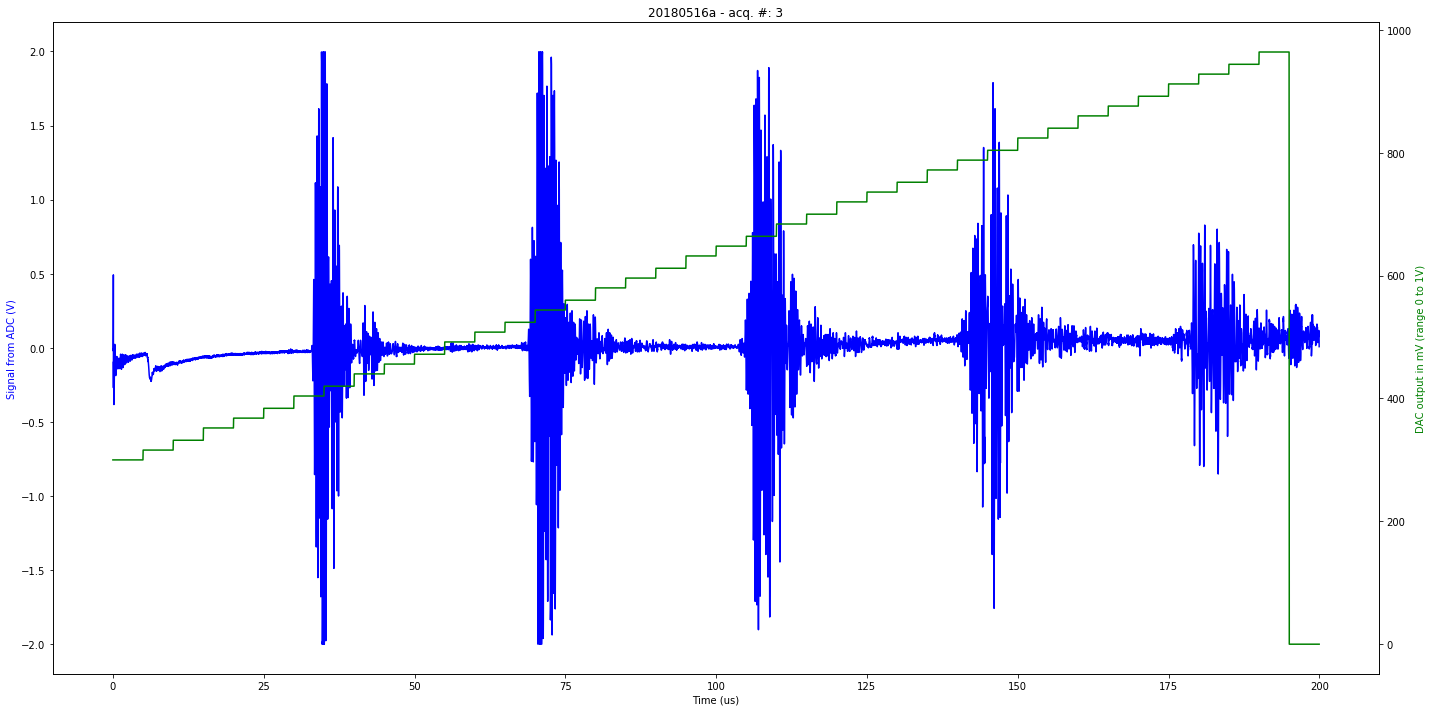
Saved at data/20180516a-3.npz
1
data/20180516a-0.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
second
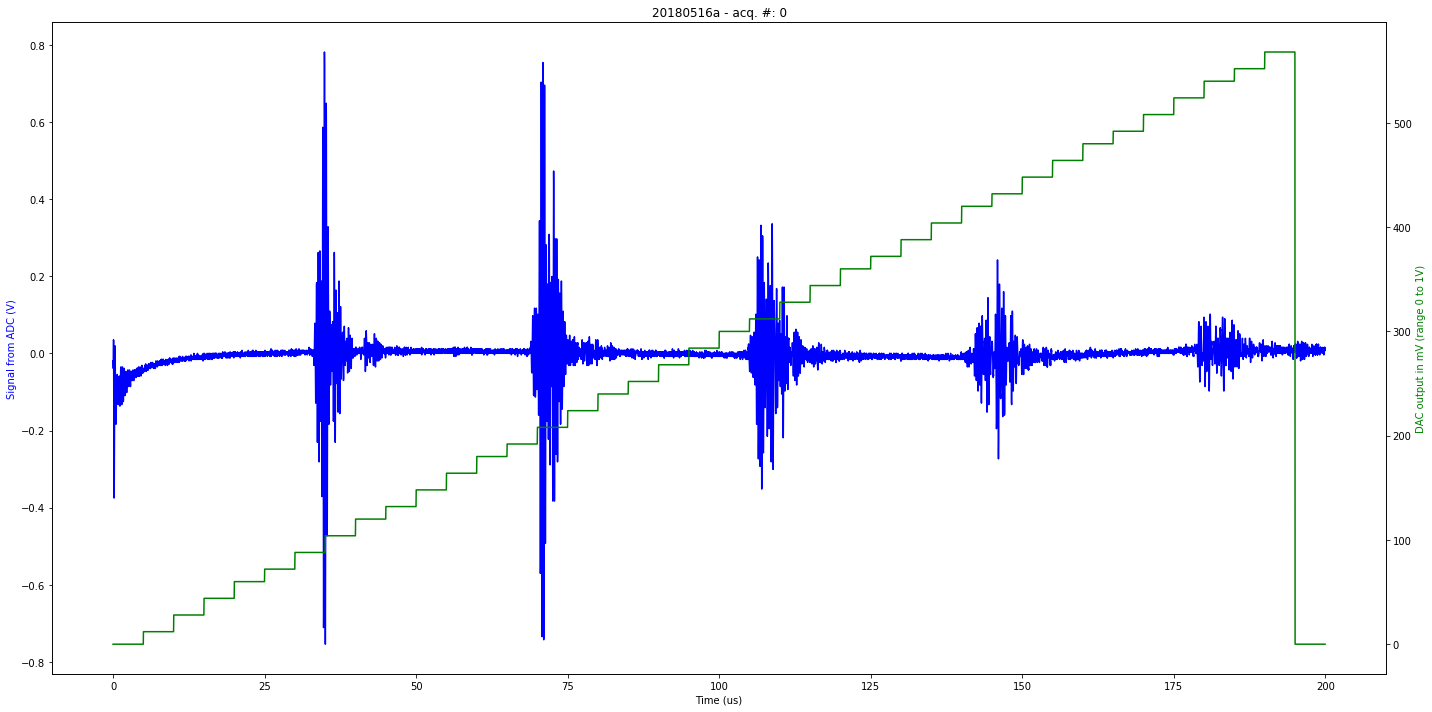
Saved at data/20180516a-0.npz
1
data/20180516a-1.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
first
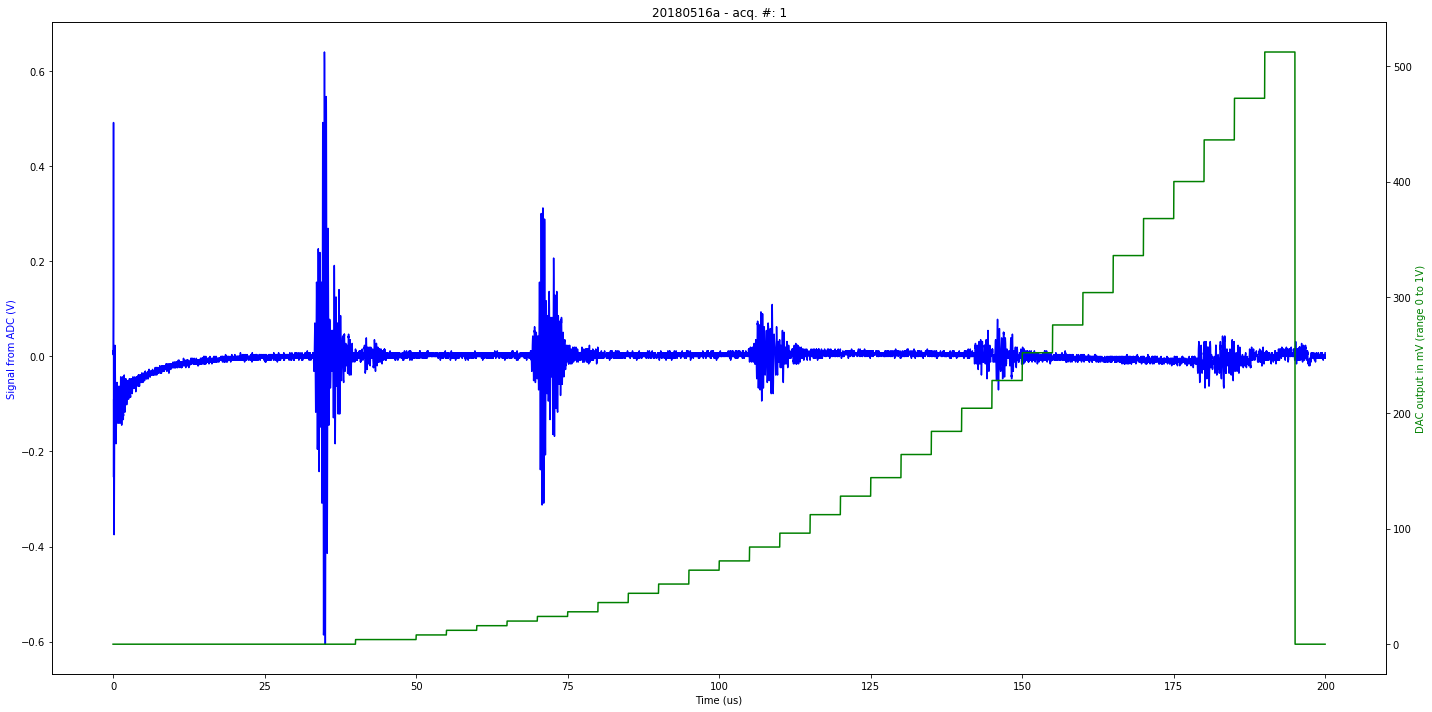
Saved at data/20180516a-1.npz
1
data/20180516a-6.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
second
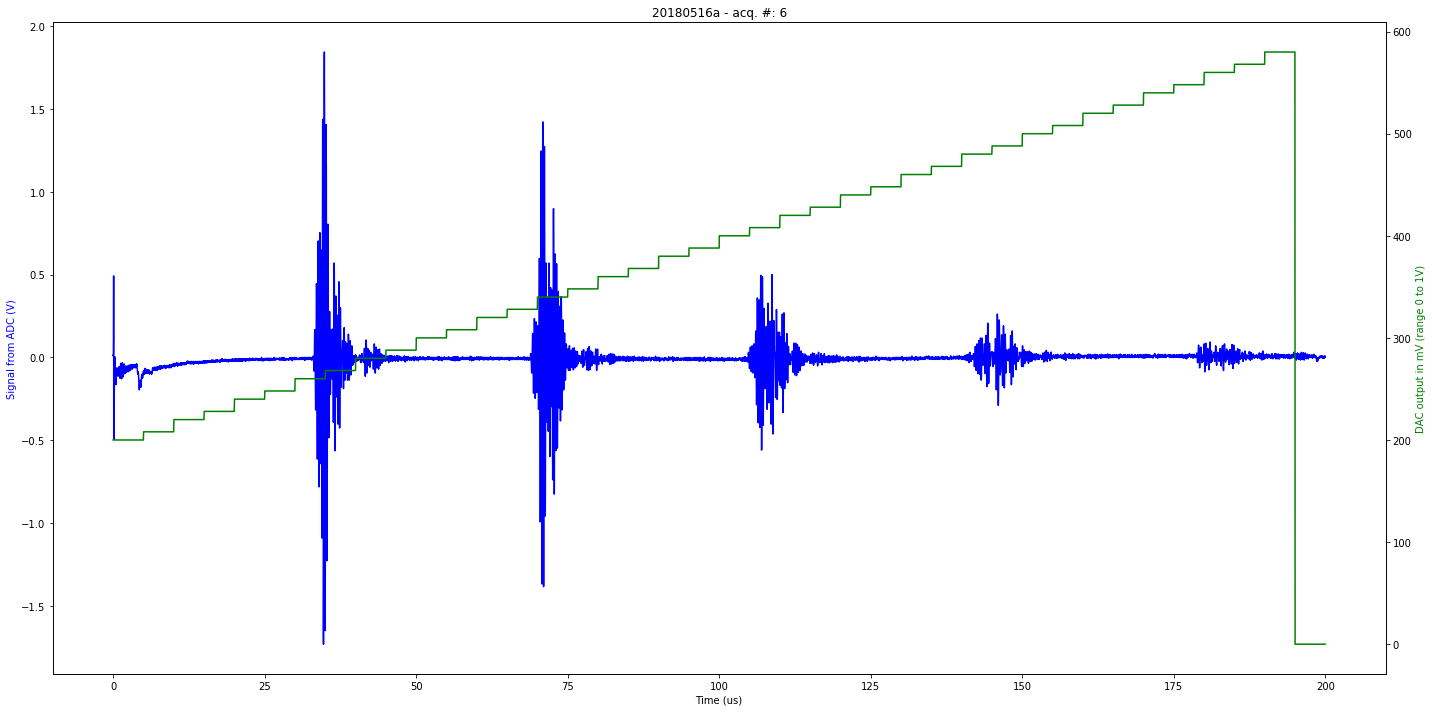
Saved at data/20180516a-6.npz
1
data/20180516a-2.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
second
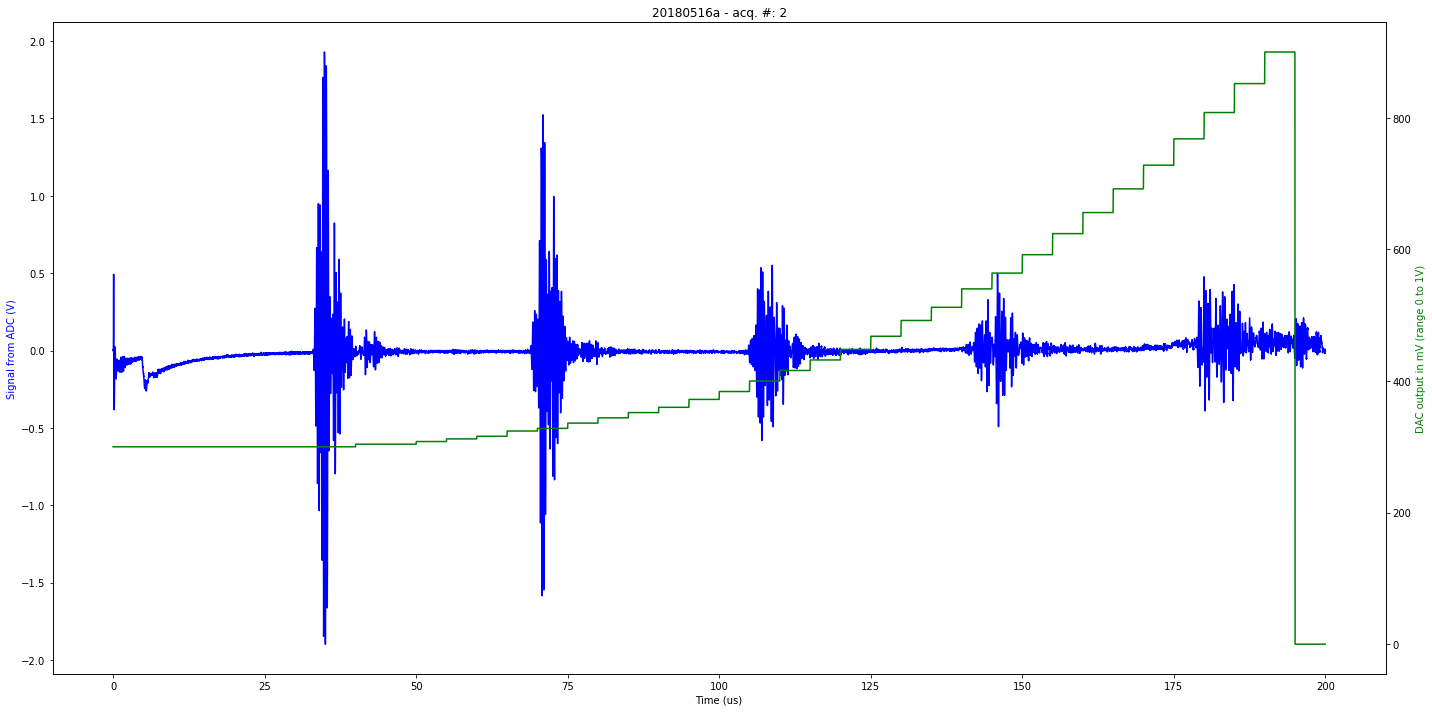
Saved at data/20180516a-2.npz
1
data/20180516a-5.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
first
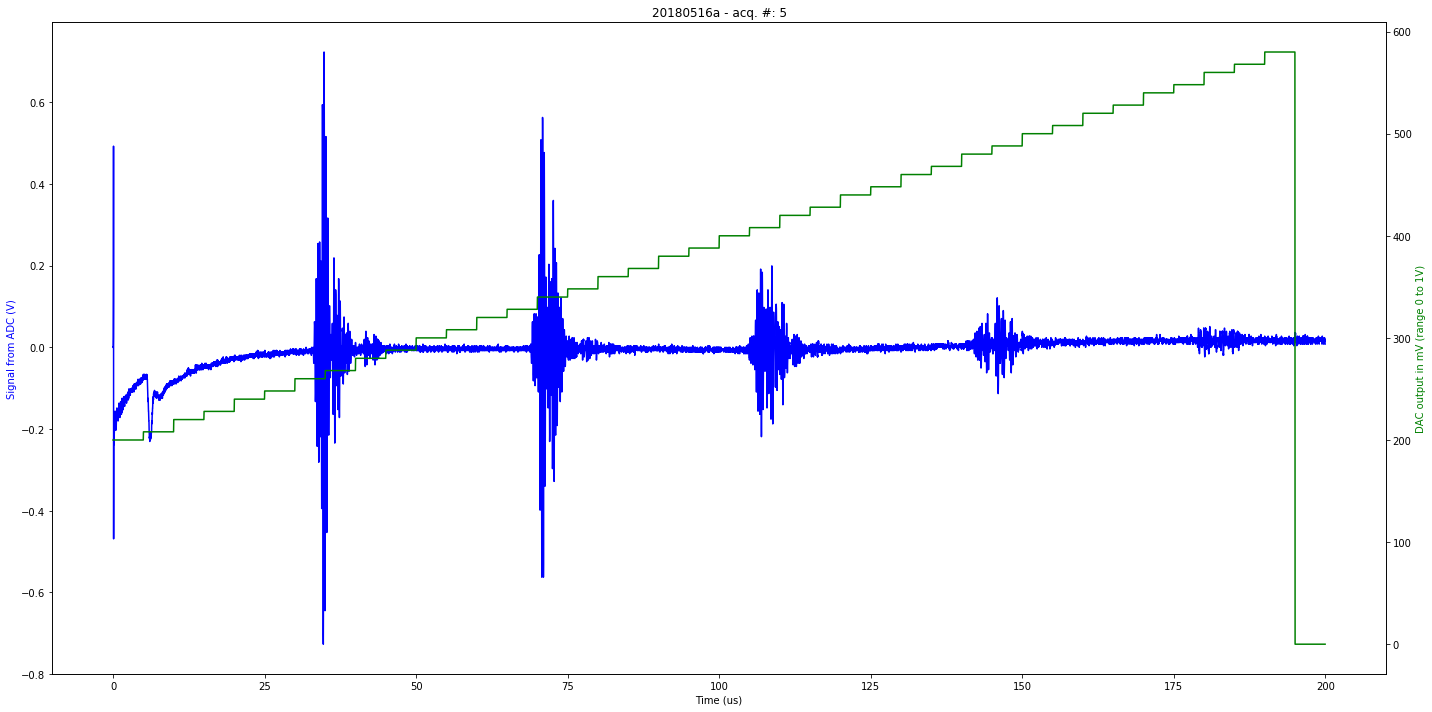
Saved at data/20180516a-5.npz
1
data/20180516a-4.json
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
second
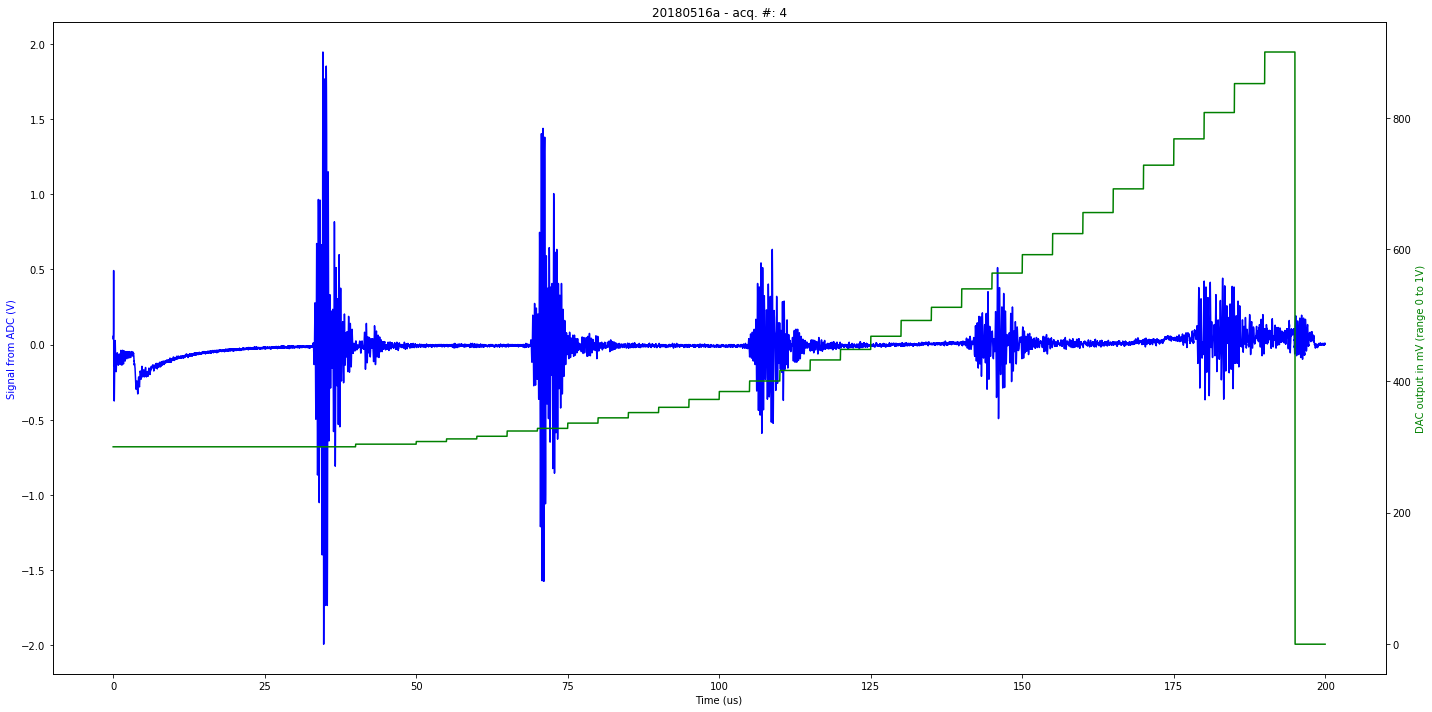
Saved at data/20180516a-4.npz
1
x = us_json()
x.JSONprocessing("data/20180516a-3.json")
x.mkImg()
x.SaveNPZ()
[u'firmware_md5', u'experiment', u'parameters', u'N', u'registers', u'V', u'firmware_version', u'data']
first
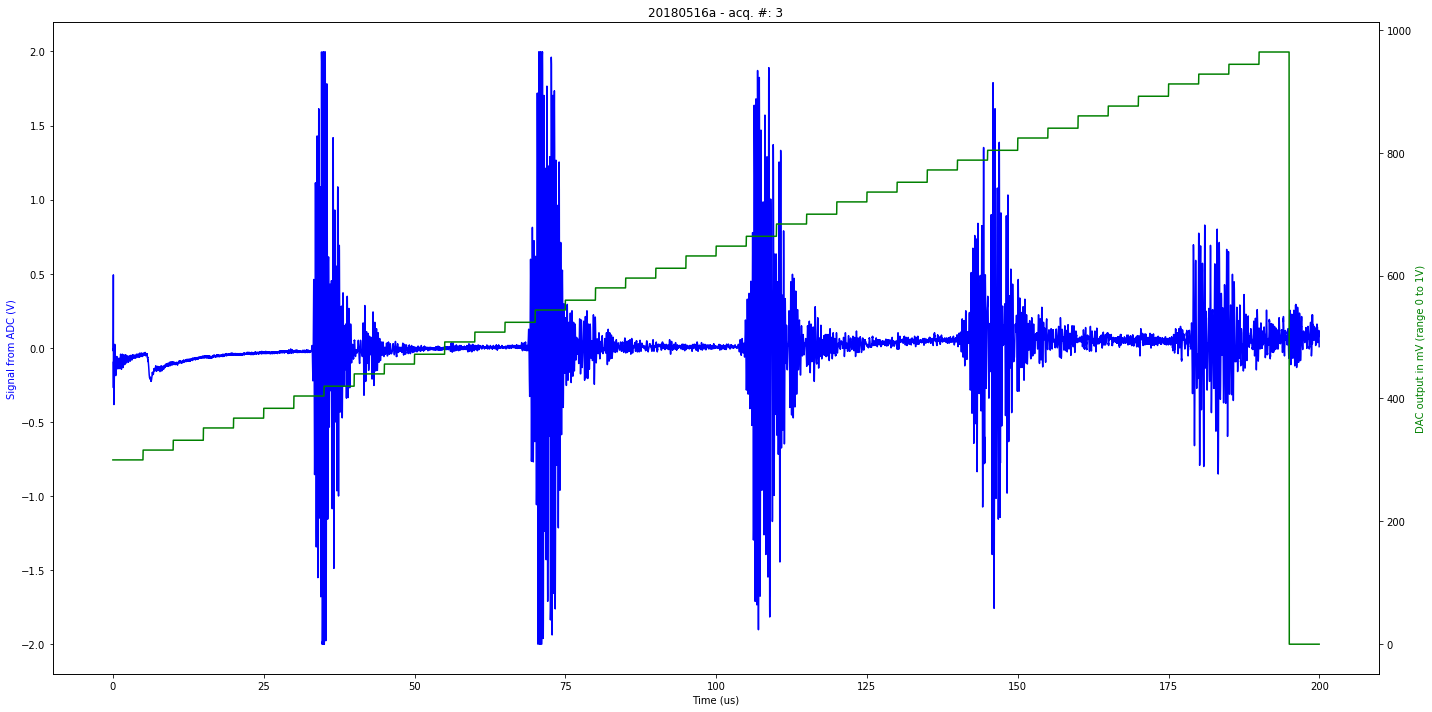
Saved at data/20180516a-3.npz
x.mkFFT()
plt.figure(figsize=(15,5))
plt.plot(x.t[12799*178/200:12799*190/200], x.tmp[12799*178/200:12799*190/200], 'b-')
plt.plot(x.t[12799*178/200:12799*190/200], x.SignalFiltered[12799*178/200:12799*190/200], 'y-')
plt.title( "FFT of "+x.iD + " - acq. #: "+ str(x.N))
plt.xlabel('Freq (MHz)')
plt.tight_layout()
plt.show()
Processing the data
Checking IDLines
plt.plot(x.t,x.IDLine)
print x.IDLine[0],x.IDLine[-1]
plt.show()
8 14














